Peering into the genomes of the tricky RTB crops
Cutting-edge tools are helping shorten expensive breeding programs. New kinds of marker-assisted selection allow breeders to see the genes of plants before growing them. Across the RTB Program, breeders are using new genomics tools to enhance the breeding of tomorrow’s crops.
RTB crops are especially difficult to breed. For example, potatoes, sweetpotatoes, yams and bananas have polyploidy (more than two sets of chromosomes — often from slightly different ancestors), giving them complex genetic structures and making them hard to breed. As a further complication, there are multiple species of banana, yam and potato. As these crops are vegetatively propagated, they are slower to breed and to bulk up seed.
Genomics-assisted breeding can help to overcome these challenges. Luis Augusto Becerra, FP 1 leader, noted that “genetic tools make it possible for breeders to see into the genetic makeup of a plant before growing it in the field. This helps us to save time and money, and it accelerates breeding.”
Therefore, across the RTB Program, scientists and breeders are developing tools so that genomics-assisted breeding can be implemented in the various breeding programs. Michael Friedmann, RTB science officer, explained, “RTB scientists are making great progress in developing genomic tools across the RTB Program. RTB is bringing together scientists and breeders to create synergies, develop new genetic methods, and enhance the efficiency of crop breeding.” Friedmann recently coordinated the publication of a study in a special edition of the journal Agriculture, which tracked this progress in using the tools across the RTB Program.
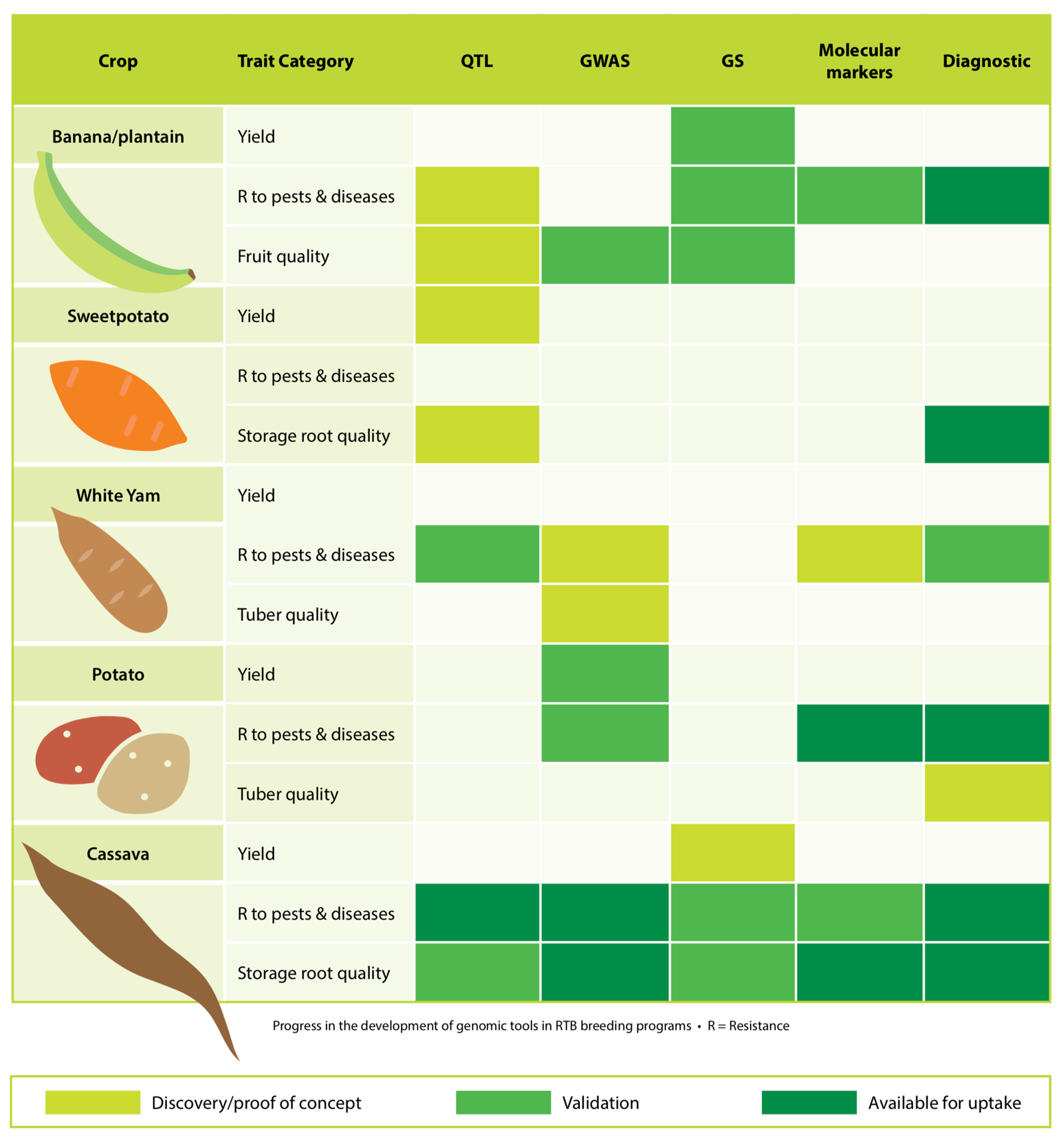
Two of the most important new tools are genome-wide association studies (GWAS) and genomic selection (GS). GWAS is a form of marker-assisted selection in which thousands of genetic markers spread across the genome are used to identify molecular markers close to genes that are associated with desired traits, with a view towards accelerated breeding. Therefore, scientists across the RTB Program have been taking the steps necessary to develop the genomic resources to carry out GWAS and GS. They have participated in the sequencing of reference genomes for all the crops. Populations with individual plants differing in the levels of important traits have been developed, their DNA sequenced and the relevant traits measured. This information has been used to carry out GWAS to tag complex traits with molecular markers for cassava, banana, sweetpotato, potato and yam. Sets of markers are now being characterized for many important traits, ranging from higher yield, biotic and abiotic stress tolerances and quality traits. For example, GWAS was applied to evaluate the existence of cassava green mite (CGM) resistance alleles in 845 advanced breeding lines. The study detected 35 genetic markers in the cassava genome. Seventeen candidate genes were found to be associated with CGM resistance, potential sources to develop pest-resistant cassava.
GS does not rely on individual molecular markers to select for traits, but rather uses the genome sequence information of an individual to predict the traits, by first developing predictive models on a training population that is both sequenced and characterized for the traits of interest. Then, new individuals are selected for a trait such as yield, based on their sequence information, that is fed into the models. RTB scientists have begun to develop and characterize these training populations and are exploring different models and strategies for the best predictive values. Consequently, models with high predictive values for banana fruit filling and fruit bunch traits were identified, showing the potential of genomic prediction to increase selection efficiency in banana breeding. Likewise, GS studies in cassava were used in Uganda to predict tolerance to cassava brown streak disease (CBSD) for 35 West African clones, based only on their sequence information. This was particularly important, since this severe disease has not reached West Africa yet, and so, breeding material in West Africa cannot be screened for tolerance to the virus.
Therefore, genomic tools can aid in the selection of superior genotypes, especially in relation to complex traits, which will result in increased genetic gains in the breeding programs. As RTB crops share complex genomes, advances in one crop can help inform strategies for the other crops. By bringing together breeders and molecular biologists, RTB is facilitating the implementation of genomics-assisted breeding across its target crops. By developing an overall framework for tracking progress in the use of these tools we can guide their uptake and use.
SHARE THIS
